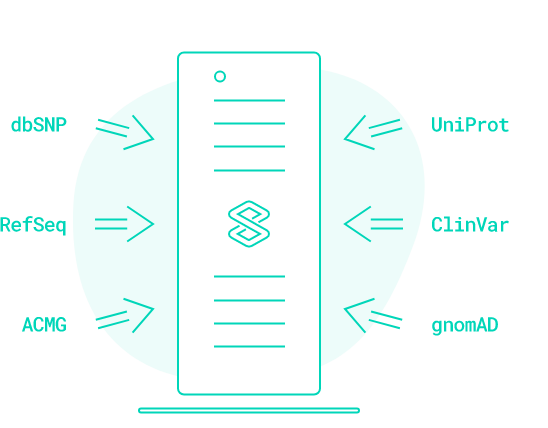
End-to-End Bioinformatics Solution for Next-Generation Sequencing

VarSome Clinical is a CE IVD-certified and HIPAA-compliant platform allowing fast and accurate variant discovery, annotation, and interpretation of NGS data for whole genomes, exomes, and gene panels.
Get in touch for a quotation